Genetics of Dravet Syndrome
Learn more about the genetic cause and biology behind Dravet syndrome
Genetics of Dravet Syndrome
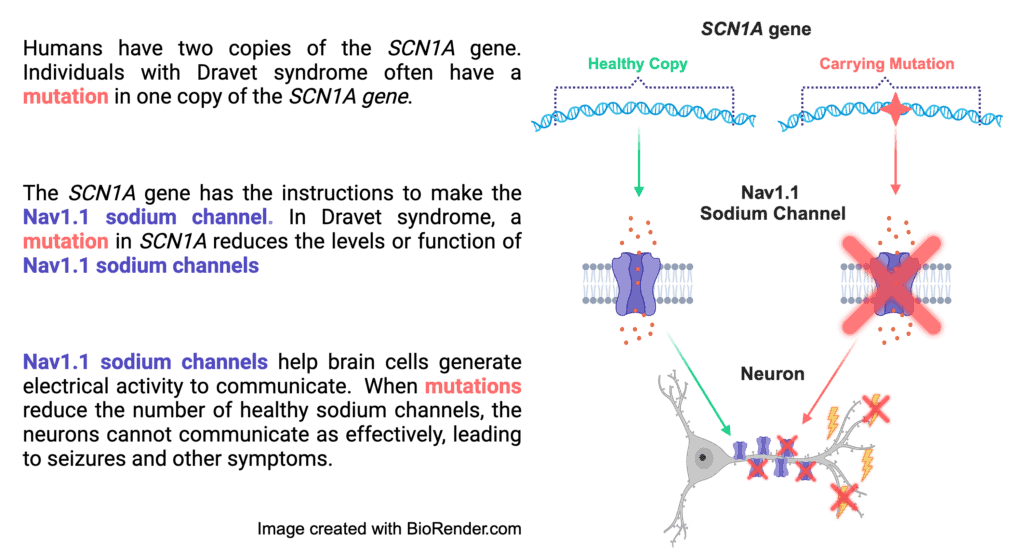
The majority of cases of Dravet syndrome are caused by loss-of-function mutations in SCN1A
More than 90% of patients diagnosed with Dravet syndrome have an SCN1A mutation. The SCN1A gene codes for the production of sodium ion channels, which are pore-like proteins embedded in the cell membrane that allow sodium ions into and out of the cell, propagating electrical signals. The mutations in SCN1A that cause Dravet syndrome lead to loss-of-function of one copy of the gene; this means half of the sodium channels either are not made or have a significantly decreased function, depending on the specific mutation. This results in a condition called haploinsufficiency, which means that one functioning copy is not sufficient to prevent symptoms.
- Approximately 90% of SCN1A mutations that cause Dravet syndrome are de novo, meaning they are not inherited from a parent. In these cases, the mutation happened randomly, and it was not passed on or ‘carried’ in a parent’s genetics.
- In about 4-10% of cases, SCN1A mutations are inherited from a parent. In these cases, there is a 50% chance of passing the mutation on to future children.
- There are over 6,000 places for a mutation to occur on the SCN1A gene. Therefore, most patients’ mutations have not been reported in other people. Even when two individuals have the exact same SCN1A mutation, their presentation of symptoms and outcomes can be very different.
- A wide variety of SCN1A mutation types have been identified in Dravet syndrome. Mutation type does not predict the severity of symptoms or long-term outcomes.
After a positive genetic test result, consultation with a genetic counselor is recommended. The genetic counselor can help you understand your loved one’s genetic report in more detail and discuss topics like heritability. While a mutation is not necessary for diagnosis, it can help to confirm a clinical diagnosis and guide treatment.
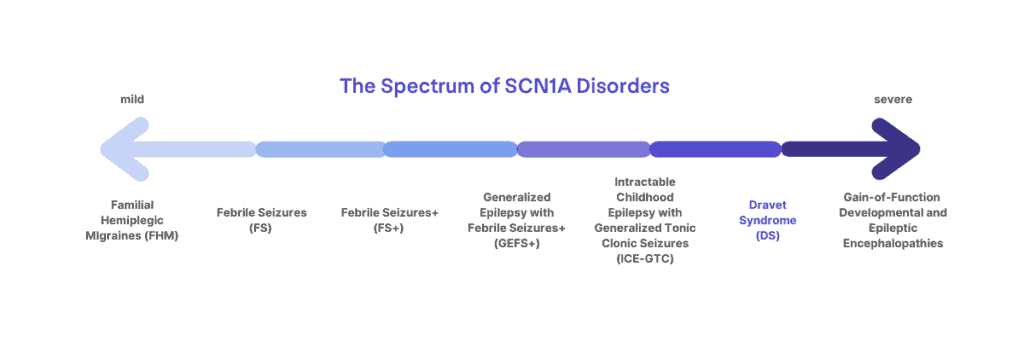
Mutations in SCN1A can cause a spectrum of disorders
Mutations in SCN1A can lead to a spectrum of disorders ranging from migraines, childhood epilepsy, or more severe and life-long epilepsy syndromes. Not every mutation in SCN1A leads to Dravet syndrome. Dravet syndrome lies at the more severe end of the spectrum of SCN1A-related disorders.
Rarely, Dravet syndrome is associated with mutations in other genes
Although rare, patients with mutations in other genes (such as GABRG2, GABRA1, SCN1B) may also be diagnosed with Dravet syndrome. In the past, patients with mutations in genes like SCN2A, STXBP1, and PCDH19 may have received a diagnosis of Dravet syndrome, but these are now grouped as unique genetic syndromes.
Gain-of-function mutations in SCN1A are associated with a unique diagnosis that may benefit from a different treatment course
Dravet syndrome is caused by SCN1A mutations that lead to loss-of-function, but recently gain-of-function SCN1A mutations have also been identified that lead to another developmental and epileptic encephalopathy with some unique features from Dravet syndrome. Gain-of-function mutations are still being studied, and may not always be identified on genetic reports. Identification is important, as patients with gain-of-function mutations may actually benefit from anti-seizure medications that block sodium channel function, while these are contraindicated in Dravet syndrome caused by loss-of-function. Gain-of-function mutations may be suspected in a patient with a non-truncating SCN1A mutation and warrant further investigation if:
- seizure onset occurs before 3 months of age
- patients present during early development with a non-seizure movement disorder
- sodium channel blockers do not worsen seizures
Any presentation of symptoms considered atypical for Dravet syndrome could be reason to look at a patient’s genetic report again with a geneticist or genetic counselor. Knowledge of human genetics is changing rapidly as we learn more about individual disease-causing variations in DNA. Patients with gain-of-function mutations have only recently been identified, and understanding the full spectrum of patients and how symptoms present is still evolving.
Frequently Asked Questions about Genetics and Dravet Syndrome
My infant has received genetic test results indicating an SCN1A mutation. What does that mean?
In years past, a mutation in the SCN1A gene has confirmed a diagnosis of Dravet syndrome in patients that were already showing the typical disease progression. When access to genetic testing was more limited, most diagnoses occurred after associated health issues had already started to develop. In more recent years, with greater access to genetic testing, patients are now receiving confirmation of the mutation at very young ages before the typical progression of the disease becomes apparent, leading to confusion in patient families on what to expect for their child’s prognosis.
There is a spectrum of disorders associated with SCN1A mutations, ranging from mild to severe, including Familial Hemiplegic Migraines, Generalized Epilepsy with Febrile Seizures Plus (GEFS+), and Dravet syndrome. Genetic testing can help to guide diagnosis, but it must be considered in the context of clinical symptoms. For example, two patients might carry the exact same mutation and still have different diagnoses, such as Dravet syndrome and GEFS+. Additionally, even within the diagnosis of Dravet syndrome, there can be a lot of variability in when and to what extent symptoms present. Published studies of the patient population can report common trends, but there are outliers and a lot of variability. Mutation type or location is not enough to determine how severe a patient’s outcomes may be or to predict things like medication responses. Research is ongoing to better understand the spectrum of patients with Dravet syndrome and other SCN1A-related disorders.
When will I know if my child has Dravet syndrome or a different SCN1A disorder?
With earlier genetic diagnoses, we will gain a better understanding of the full spectrum of SCN1A disorders in the coming years. Until then, once seizures occur and a mutation in SCN1A is identified, it is essential to follow a treatment protocol based on the international treatment consensus for Dravet syndrome*. This ensures that contraindicated medications are avoided and that patients receive the highest quality of care. Earlier intervention with the most appropriate treatment plans may also improve the overall picture of long-term outcomes for patients with Dravet syndrome. *In a small proportion of cases, mutations in SCN1A may be Gain-of-Function, rather than the Loss-of-Function mutations that lead to Dravet syndrome. Patients with Gain-of-Function mutations in SCN1A may respond to a different treatment protocol. Some of the key clinical signs that may suggest looking to see if there is a Gain-of-Function mutation including seizures onset before 3 months of age, non-seizure movement disorders, and/or joint contractures.
Some clinicians may decide early on to give an infant the diagnosis of Dravet syndrome based on the seizure types and other symptoms in conjunction with an identified mutation in SCN1A, while other clinicians may wait to see if other developmental areas are impacted before giving an official diagnosis. Regardless of the eventual final diagnosis and long-term outcomes for patients, early intervention and appropriate medication choices based on the expert-consensus recommendations give patients the best chance for positive outcomes regardless of where they land on the spectrum of SCN1A-related disorders.
One of the biological parents of my child has also tested positive with an SCN1A mutation. Does this mean that this parent also has Dravet syndrome?
The diagnosis of Dravet syndrome is based on clinical symptoms. Genetic testing can help to direct or confirm the diagnosis, but a mutation in SCN1A alone is not sufficient to automatically give someone the official diagnosis of Dravet syndrome. As mentioned above, there can be a range of disorders that result from mutations in SCN1A. Keep reading below for more explanation about how the same mutation could result in different outcomes in two individuals.
The International League Against Epilepsy helps to set the criteria for diagnosis of Dravet syndrome. You can read more on this page, but in brief, a diagnosis of Dravet syndrome involves:
- Recurrent, often prolonged seizures (often associated with fever but also unprovoked; focal or generalized)
Onset of seizures between 1-20 months (although diagnosis should be looked at more carefully if on the very early or late range of those ages) - Typical development of infant at onset with a normal EEG outside of seizures (developmental impacts begin in early childhood and slowing of the EEG is common over time)
- Medication-resistant seizures that may evolve over time
- Intellectual disability that becomes apparent generally by school age (commonly moderate to severe; although there are rare reports of individuals who are less impacted in this area). The majority of patients with Dravet syndrome will need supportive care throughout their lifetime.
- Additional symptoms commonly associated with Dravet syndrome include speech delays or impairments, behavior disorders, gait and movements disruptions (such as crouched gait developing during adolescence)
How is it possible for both my child and I to have an SCN1A mutation, yet I don’t have Dravet syndrome?
In a small portion of the patients with Dravet syndrome, mutations in SCN1A have been inherited from a parent who carries the mutation. On the surface this can be confusing, why did the mutation cause Dravet syndrome in the child but did not cause the parent to have the symptoms and diagnosis of Dravet syndrome?
There are actually several factors that can impact how someone’s genotype (the actual ‘code’ of their DNA) impacts their phenotype (the physical symptoms that present from their DNA ‘code’). It is not unique to Dravet syndrome, or the SCN1A gene, to have variability in the presentation of symptoms from the same or similar mutations. Factors that can impact the phenotype resulting from a mutation include:
- Other parts of the person’s genetic code; for example, another small gene change that may compensate for a significant mutation in another gene. Studies of animal models and families with inherited SCN1A mutations have shown that there can be ‘modifier genes’ or additional mutations that are compensatory which impact the severity of the phenotype from an SCN1A mutation.
- Environmental factors or exposures. In the context of Dravet, this could include things like being on contraindicated medications for long periods of time. In animal models, early-life stressors have been shown to worsen long-term outcomes.
So a mutation in SCN1A alone is not enough to always say “Dravet.” Which is why phenotype (or clinical symptoms) have to be taken into account with genotype (the mutation in SCN1A) to determine an accurate diagnosis, since there are scenarios where individuals carry a mutation that could be causative but do not develop symptoms (or as severe of symptoms).
Do healthy siblings or other family members need to be concerned about passing on Dravet syndrome?
In the majority of situations (90%), the mutations that cause Dravet syndrome are not inherited from parents, meaning there would not be any impact for unaffected siblings because other family members are not ‘carrying’ the gene mutation. However, in the situation where the genetic mutation has been inherited from a parent, siblings may want to speak with a genetic counselor and seek genetic testing. Often in families where genetic mutations causing Dravet syndrome are inherited, there is a family or parental history of seizures or other less severe form of epilepsy.
Genetics and Mutation Types
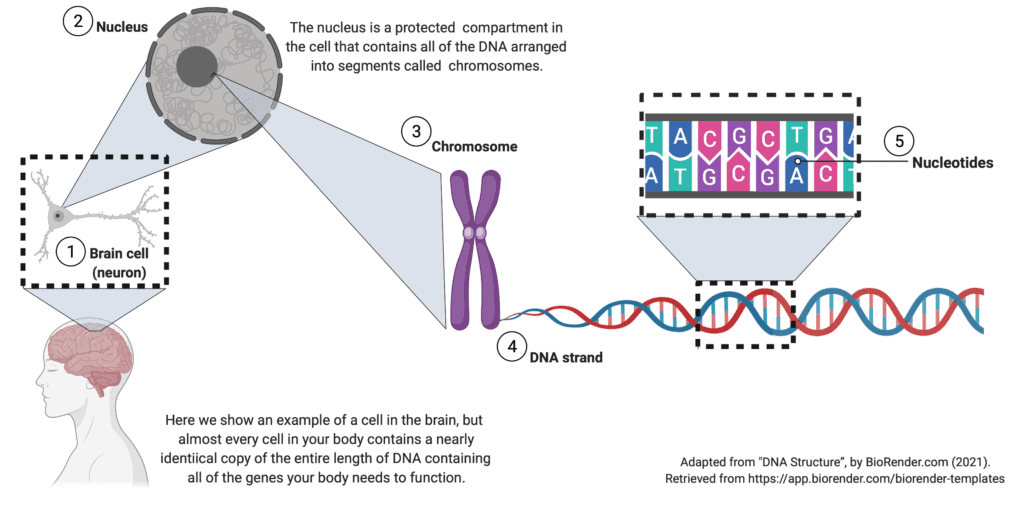
What is DNA?
DNA is the set of instructions contained within each of our body’s cells. The instructions tell the cell how to build the proteins it needs to function. A strand of DNA is a long chain of 4 different nucleotides (abbreviated A, T, C, and G) strung together in a particular order, billions of nucleotides long. Because there is so much DNA in our cells, it is organized into 23 pairs of chromosomes, much like two sets of encyclopedias would be organized into 23 volumes each. When a sperm and an egg, each containing 23 chromosomes, combine, the result is 46 total chromosomes, organized into 23 matching pairs.
What is a gene?
The 23 pairs of chromosomes are further divided into smaller segments called genes. A gene is much like a chapter in an encyclopedia and contains the instructions for producing a specific protein. Each gene is a small segment of DNA and is thus also a long chain of 4 different nucleotides (A, T, C, and G) strung together in a particular order. Because our cells have one copy of each gene from each parent, every cell has two copies of each gene unless the gene is carried on the sex-determining X or Y chromosome. SCN1A is not on the X or Y chromosome.
Genes are read in groups of 3 nucleotides called codons. Each codon is translated into one of twenty amino acids, which are then strung together like beads on a necklace. The amino acids interact with each other based on their chemical properties similar to how magnetic beads attract and repel each other when folded up in one’s hand. As the amino acids interact, the long chain folds on itself to form a very specific 3-D shape.
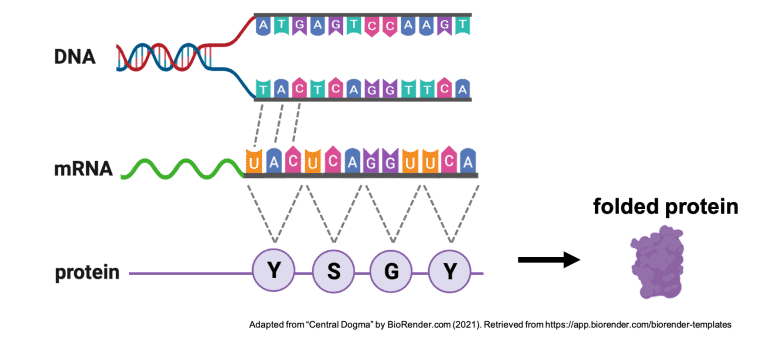
In the case of SCN1A, this 3-D shape is an ion channel, called Nav1.1, that functions as a gated channel in the cell membrane, letting sodium ions into and out of the cell. The inward and outward flow of ions like sodium allows electrical signals to be generated along neurons. These electrical signals are how the cells in our brain, neurons, communicate with each other and the rest of the body.
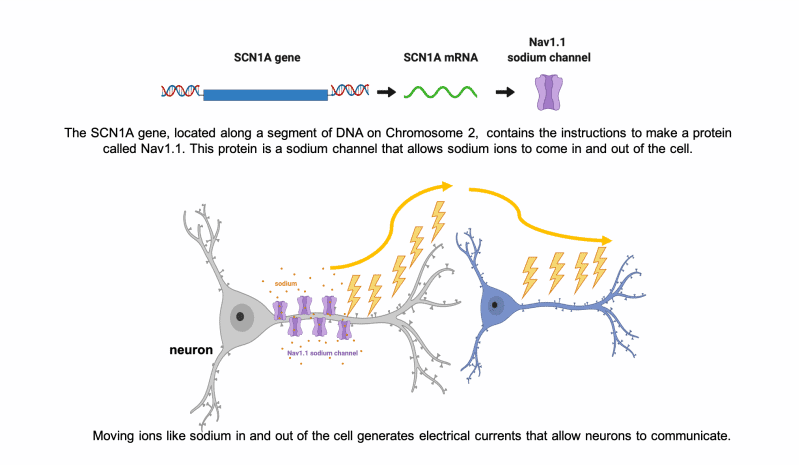
What is a mutation?
A mutation is a change in the expected sequence of nucleotides (the order of the ‘A’,’T’,’G’,and ‘C’ units) within a gene. This change in the original sequence of DNA may then alter the sequence of amino acids that are translated from the DNA code to build the protein. A mutation may cause the chain of the protein to end too early, fold incorrectly, or otherwise change the ability of the sodium ion channel to function properly. Often the mutations that cause Dravet syndrome severely impact the protein, so that it is either never made or is broken down by the cell. Dysfunctional sodium ion channels can result in improper electrical activity and seizures.
SCN1A is a very large gene, made up of about 160,000 individual nucleotides (A, T, C, and G’s). Although SCN1A has 160,000 nucleotides, the body edits this sequence of 160,000 down to about 6,000 in the final SCN1A mRNA transcript that serves as the instructions for making the sodium ion channel [http://ghr.nlm.nih.gov/gene/SCN1A]. Still, with over 6,000 nucleotide positions, it is no wonder that most mutations reported in the literature have not yet been seen in another patient.
Remember that every cell actually contains two copies of SCN1A; one from each parent. Usually, only one copy is mutated, a condition termed heterozygosity [Brunklaus et al 2015]. Approximately 4% of the mutations seen in Dravet syndrome are inherited directly from parents, with the parent often experiencing fewer and less severe symptoms than the child in a phenomenon known as reduced penetrance [Xu et al 2012]. This could be because of other impacts of their personal genetic make-up, or something called mosaicism (discussed more in a section below).
SNP’s:
Mutations are actually a natural phenomenon that has been occurring in all organisms for thousands of years. Most changes in DNA sequence have little to no effect on the final protein products because they occur in regions that are edited out during gene processing, or their location in the final protein does not alter its function. In fact, many members of the healthy population have variants in their genes that are shared with a significant percentage of the population. Because these variants have no obvious clinical symptoms, they are called single nucleotide polymorphisms (SNP’s) and are not considered mutations. Your lab report may include these SNP’s, but their presence is not considered a positive SCN1A test.
Mutation Types
There are several types of mutations that can occur in the SCN1A gene and result in Dravet syndrome. Read on to learn about each mutation type, including some tips that may help in understanding a genetic report.
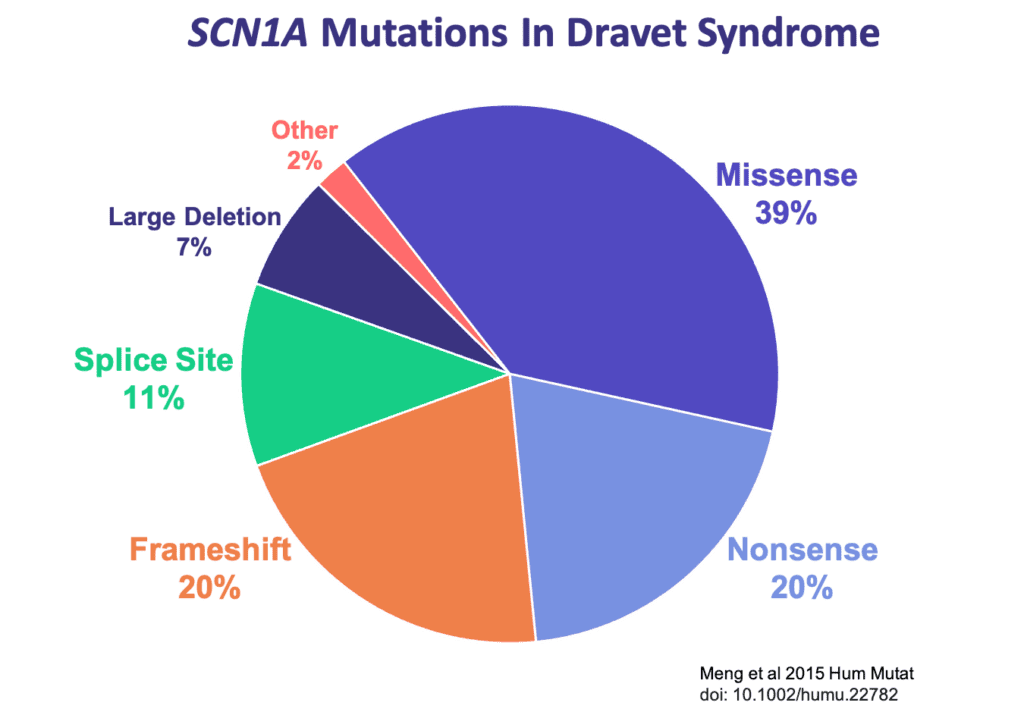
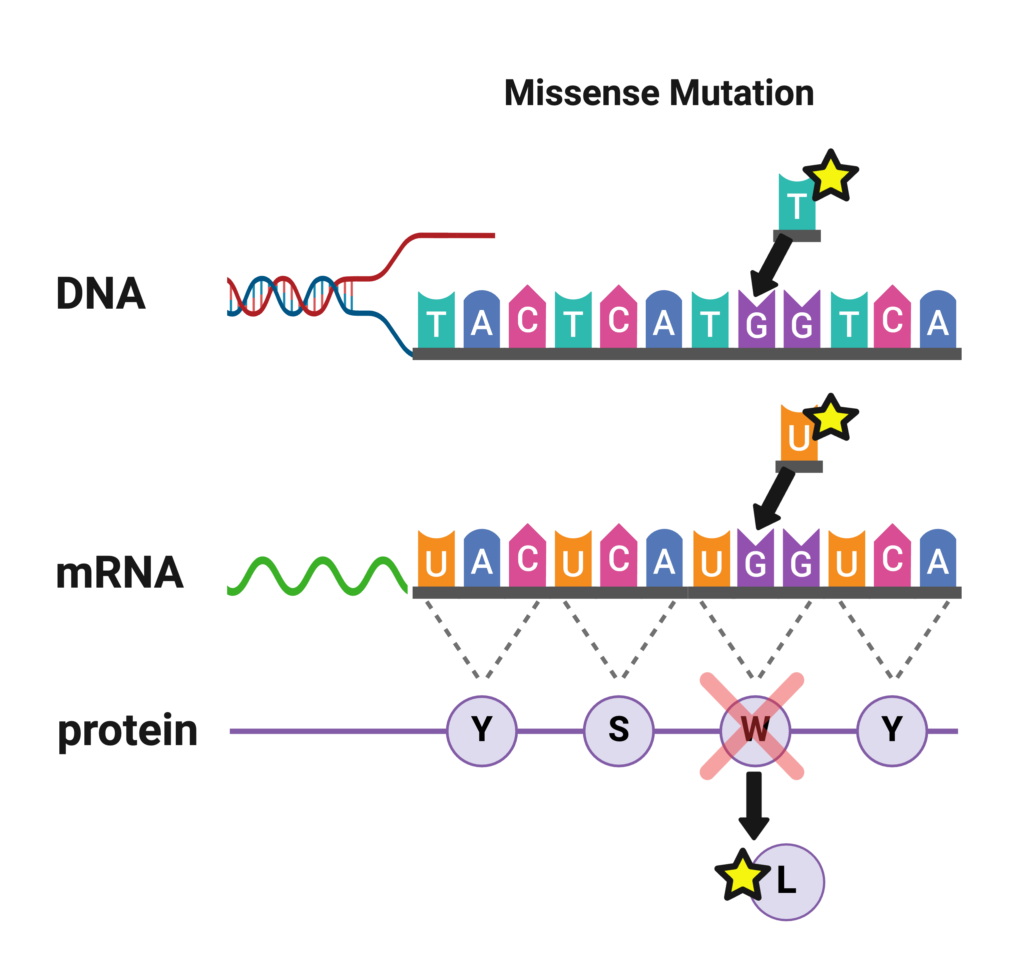
Missense Mutations
Missense mutations are the most commonly identified mutations in patients with Dravet syndrome. A missense mutation is a simple substitution of one nucleotide for another at a single location in a gene. This slight change in the sequence of nucleotides alters the code for an amino acid in the long chain that forms the protein, which can impact the ability of the protein to function. Depending on the type and location of the change in the code in SCN1A, there may be differing impacts on how severely the Nav1.1 sodium channel is affected.
If a missense mutation occurs near a pore-forming region of the sodium ion channel, it is likely to significantly alter the ion channel’s function and cause a more severe case of SCN1A-related epilepsy such as Dravet syndrome [La Selva et al 2003]. If a missense mutation occurs at a less critical location on the gene, it may produce milder clinical symptoms or no symptoms at all. Approximately 39% of the mutations seen in Dravet patients are missense mutations [Meng et al 2015].
A missense mutation reported by a testing company may look like this:
- Variant 1: Transversion G>T
- Nucleotide Position: 4073
- Codon Position: 1358
- Amino Acid Change: Tryptophan>Leucine
- Variant of Unknown Significance (heterozygous)
The same information could also be reported as: SCN1A c.4073G>T, p.Trp1358Leu (W1358L).
This all means that the mutation in the gene was a substitution of T in place G at the 4,073rd nucleotide position (out of 6000 in the final gene that is read). Remember that the nucleotides that make up DNA are ‘read’ in groups of 3, called codons, so 4073 divided by 3 gives you the amino acid or codon position of 1,358. The amino acid at this position, called Leucine (abbreviated as ‘Leu’ or ‘L’) was substituted for the amino acid Tryptophan (abbreviated as ‘Trp’ or ‘W’).
In this example, the genetic testing lab reported the result as a “Variant of Uncertain Significance,” which means they were unsure based on the published information whether this mutation would lead to disease or not. If a mutation has been reported previously as associated with disease, it may be listed on the report as ‘pathogenic’. As we learn more about genetics and human disease, variants can be reclassified that were previously listed as uncertain.
The example results also reported the mutation as heterozygous, this means the patient only carries the mutation on one copy of their gene and not both, as is the case in general with Dravet syndrome.
This example of a real-life mutation is indeed located in a pore region of the sodium ion channel, and this patient does have Dravet syndrome.
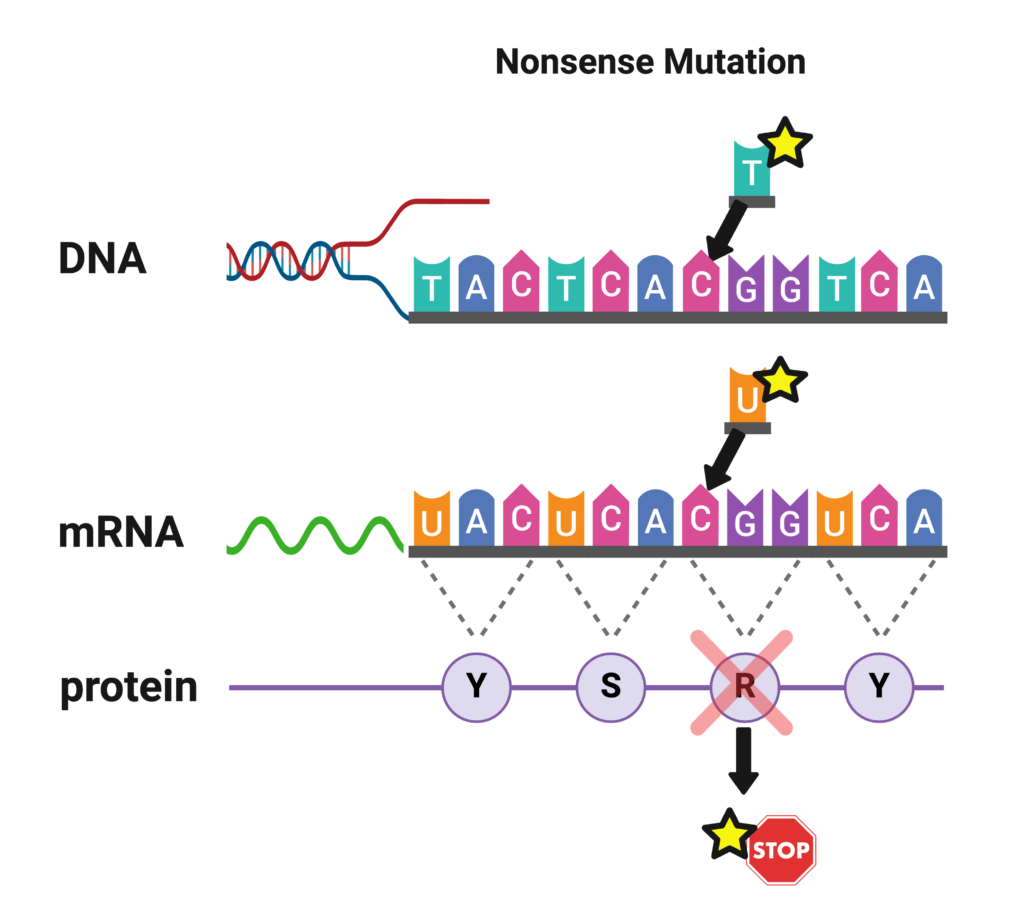
Nonsense Mutations (Truncating, Premature Stop)
Nonsense mutations are similar to missense mutations in that one nucleotide is substituted for another. However, in the case of a nonsense mutation, that substitution causes the codon to be read as a “STOP” signal. The cell stops reading the gene prematurely, and the protein is significantly shortened, or truncated. Nonsense mutations are often associated with more severe SCN1A-related epilepsies such as Dravet syndrome [Meng et al 2015]. Approximately 20% of mutations in Dravet syndrome are nonsense (truncation) mutations [Meng et al 2015]. A nonsense mutation may be reported like this:
“The mutation c.3985C>T (heterozygous) resulting in a termination or stop codon at Arg 1329 was detected in exon 20 of the patient sample and is associated with Dravet syndrome.”
This says that the nucleotide C was replaced with a T at position 3985, which resulted in the amino acid Arginine being replaced with a stop codon at position 1329. (3985 nucleotides, read in groups of 3, correspond to 1329 amino acids.) Only one of the two copies of SCN1A in the patient is mutated (heterozygous), as is usually the case. “Amber,” “Opal,” and “Ochre” may appear on the lab report and are some of the names for stop codons. The lab can be fairly confident this mutation is disease-producing because of the high correlation between truncation mutations and Dravet syndrome.
This same mutation may be reported by another lab like this:

This report does not specify the nucleotide position, but it identifies the amino acid position as 1329, and the asterisk (*) next to the amino acid position indicates a stop codon. Again, the lab is confident this mutation will result in Dravet syndrome.
Frameshift Mutations (Insertions/Deletions)
Sometimes, one or more nucleotides are deleted from the gene. Because the DNA code is ‘read’ in sections of three nucleotides at a time, if one or two nucleotides are inserted or deleted, the reading frame of codons is shifted, and every amino acid is incorrect from that point in the chain on. This usually causes a dysfunctional sodium ion channel. In addition, the shift in reading frame will often cause one of the codons farther down the chain to be interpreted as a stop codon, prematurely terminating an already dysfunctional chain.
If a group of 3 nucleotides is inserted or deleted, only one codon is added or deleted, respectively, and the protein may still be functional depending on the location of that insertion or deletion.
On a genetic report, frameshifts may be indicated by the letters “fs” following the affected amino acid. If the frame shift leads to an early stop, or termination, it may be listed as “fs Ter#”, with the number referring to how far along the chain the stop codon is located following the shift.
Splice Site Mutations
Genes contain the ‘code’ for proteins- in the case of SCN1A it holds the ‘code’ for the sodium channel NaV1.1. The DNA around a gene contains additional information that is not necessary to make the protein, so cells use something called “splicing” to cut out the extra information in the DNA down into a shorter message (called RNA) that just has the information that is needed to make the protein. In rare situations, mutations can occur in the sections of the DNA that impact how a gene is spliced, resulting in a major disruption to that copy of the gene. These are called ‘splice site mutations.’
Large Deletions
A small proportion of individuals with Dravet syndrome have larger deletions in their DNA. These might remove a portion of one copy of the SCN1A gene, or other times, it may be an even larger deletion that removes several genes in the surrounding area of the DNA. Deletions that impact more than just SCN1A could lead to clinical presentations that may differ somewhat from a classical presentation of Dravet syndrome depending on the other genes that are impacted. Larger sized deletions account for roughly 7% of Dravet mutations [Meng et al 2015].
Mosaicism
Mosaicism:
When the mutation occurs in the sperm, egg, or very soon after fertilization, all of the daughter cells derived from the growing embryo will contain the mutation. This is the case for most mutations found in Dravet syndrome [Xu et al 2012, Meng et al 2015].
However, if the mutation occurs later in the development of the embryo, only the cells descended from the mutated cell will carry the mutation. The cells descended from the non-mutated cells of the embryo will remain healthy. This results in an individual who is mosaic for the mutation. The later the mutation took place during development of the body, the lower the percentage of cells descended from the mutated cell, and the lower the “% mosaicism” or “mosaic load.” (This is a broad generalization: In reality, the degree of specialization of the cells at the time of mutation plays a significant role in where the mutated cells are concentrated in the patient’s body and what the ultimate mosaic load is.) One study reported that SCN1A mutations with 12-25% mosaic load were potentially pathogenic, with reduced penetrance, meaning not all who carried the mutation in mosaic form exhibited signs or symptoms [Meng et al 2015].
Variants of Uncertain Significance
Sometimes a genetic test report may report an SCN1A mutation as a ‘variant of uncertain significance’ or ‘VUS’. This means that there is not enough information to know if this mutation is disease-causing (‘pathogenic’) or just a normal variation in the genetic sequence (‘benign’). Since mutations that cause Dravet syndrome can occur all across the gene, it is common to find a patient with a specific mutation that has never been reported before. Over time, as our knowledge of genetics and human disease continues to grow, variants can be reclassified.
The Biology Behind Dravet Syndrome
Our brain uses electrical currents to spread communication. These electrical currents are maintained by a balance of positive and negative charges that are carried by small charged molecules like sodium, potassium, calcium, and chloride (also called “ions”). When the cells in our brain are unable to move these ions in the correct way at the correct time, it disrupts communications. Sometimes that means neurons communicate “too much,” spreading too much electrical activity to their neighbors, which can lead to seizure activity. In many individuals with Dravet syndrome, they have a mutation that affects the ability to regulate electrical currents using sodium ions. If you want to know more, the expandable sections below provide details about how this works at the level of individual brain cells.
Neurons communicate using electrical currents.
One of the major cell types in our brains are neurons. Neurons have long extensions that form connections with other neurons so they can communicate with each other. Neurons communicate using electrical currents that travel down their long extensions to the next neuron, similar to the wires that carry electricity to our appliances.
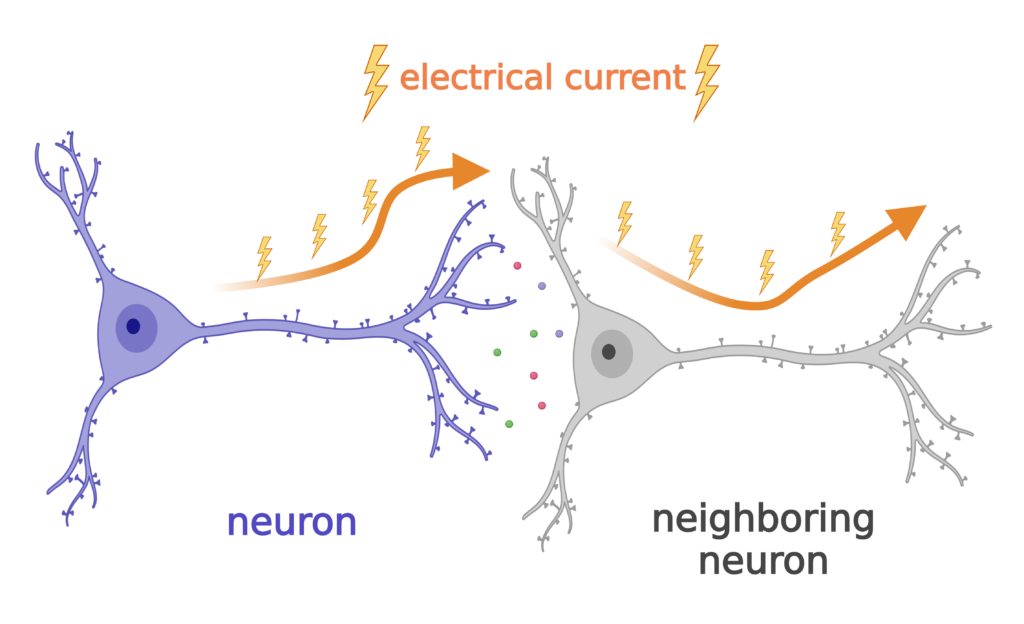
Neurons use ions like sodium to generate electric current for communication.
How do neurons generate electricity?!?
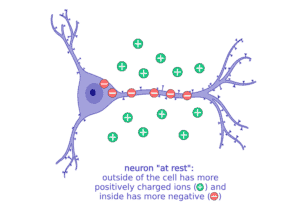
It’s all based on the movement of charged particles, or “ions,” that have positive charges (+) and negative charges (-). Sodium (Na, + charge) and potassium (K, + charge) are two important ions that neurons move to set up electrical gradients. When a neuron is “resting,” or not talking to a neighbor neuron, it is more negatively charged inside and keeps more of the positive charged ions, like sodium (Na+), outside the cell.
When a neuron gets stimulated and needs to signal with a message, it suddenly lets a bunch of the sodium (Na+) rush into the cell to start an electrical current. All of these changes in the balance of positive and negative ions leads to an electrical current that can move very quickly down the long extensions of the neuron to communicate to the next cell.
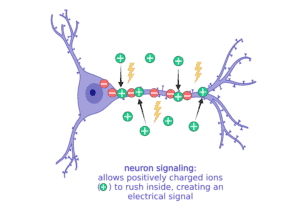
When it’s time to stop communicating, the neuron has to reverse the ions so that there are more negative inside again and more positive outside. Balancing ion movement is an important step for the cell to stop the electrical activity. This means moving more positive ions out than it brings in.
Nav1.1 is a sodium channel encoded by the SCN1A gene. Neurons use channels like Nav1.1 to move the ions that generate electric current.
To move ions inside and outside of the cell, the neuron needs special channels. There are specific channels for each type of ion (sodium, potassium, etc).
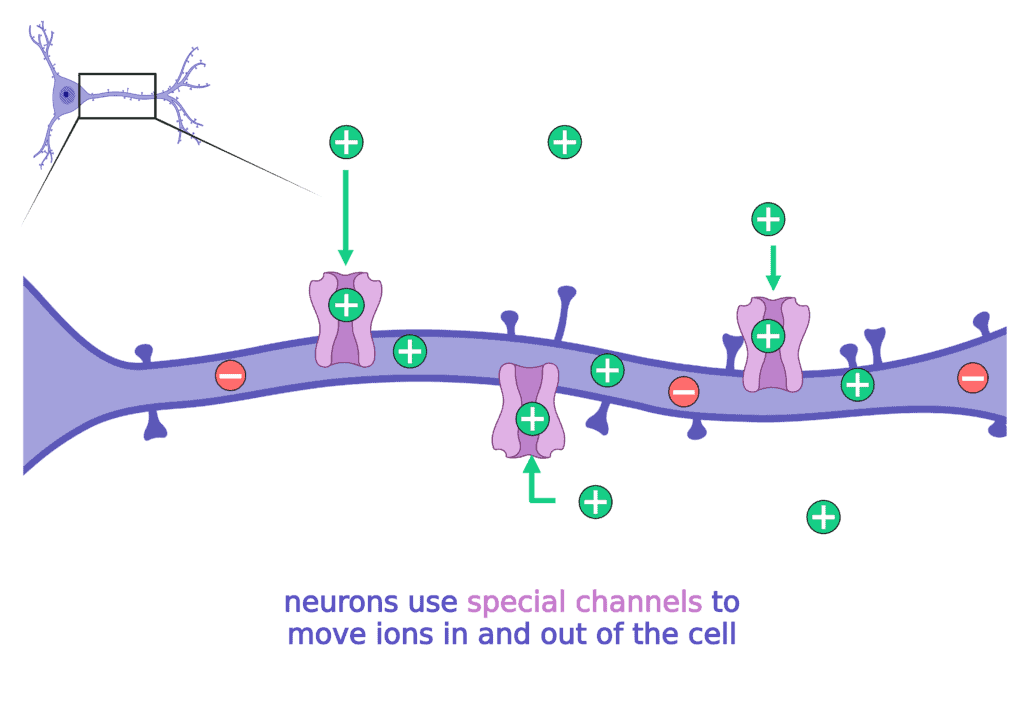
When these channels do not work properly, the neuron has a hard time making, regulating, and then stopping the electrical currents. When neurons are unable to regulate electrical currents it can lead to seizures.
The most common cause of Dravet syndrome results from a problem with a specific sodium channel, called Nav1.1.
Nav1.1 : Na for sodium + V for electrical voltage + 1.1 because there are several subtypes of sodium channels.
Most patients with Dravet syndrome have a mutation in SCN1A, the gene that has the code to make Nav1.1.
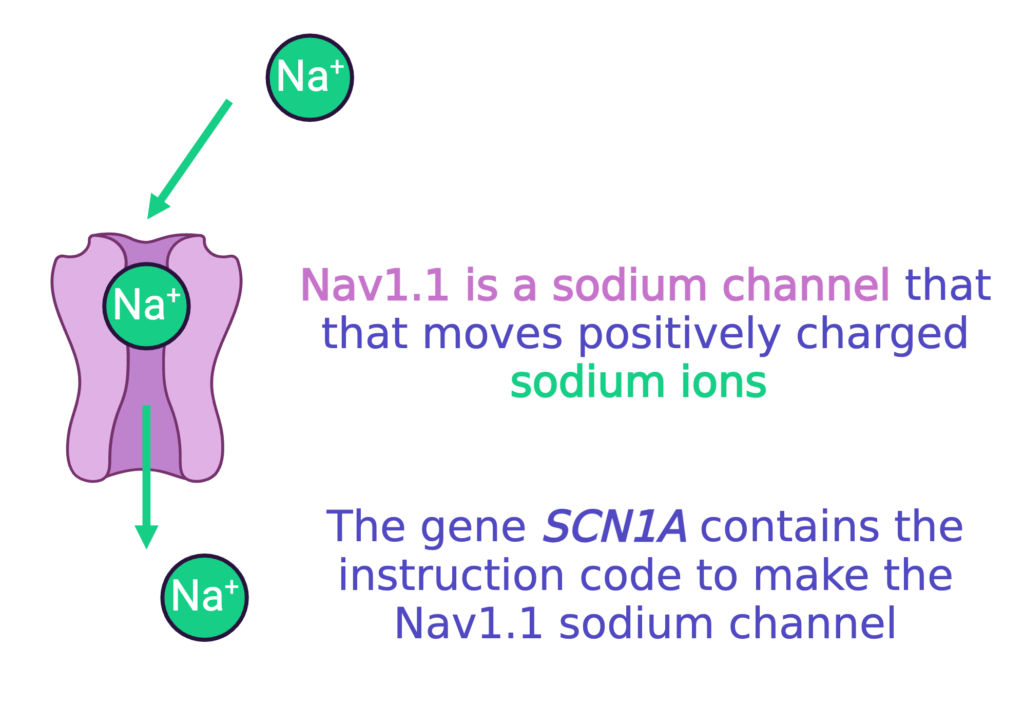
The mutations in SCN1A that cause Dravet syndrome usually cause the loss-of-function of one copy of SCN1A (we get two copies of every gene in our DNA). That means about half of the Nav1.1 sodium channels that the neuron needs either (1) do not work correctly, or (2) do not get made at all. This ultimately means less sodium channels that work correctly. This makes it very hard for neurons to properly make and stop electrical currents for communication, which can lead to seizure activity.
Reduction of the Nav1.1 sodium channel impacts the ability of specialized neurons to communicate.
This last section gets into the very specifics of what can go wrong in Dravet syndrome. At this point, you may wonder:
- if you have to bring sodium (Na+) into the neuron to start an electrical current,
- and this channel to bring sodium (Na+) into the neuron does not work as well in Dravet syndrome,
- should the problem be with too little electrical current and not too much?!
To understand how this is working to cause seizures in Dravet syndrome, we have to step back and talk about how there are different TYPES of neurons in the brain.
- When some neurons use electrical currents to talk to their neighbors, they always tell them to “get excited” and keep spreading the message. These are called excitatory neurons.
- A different group of neurons always uses their electrical currents to tell their neighbors to “STOP signaling!”. These are called “inhibitory” neurons.
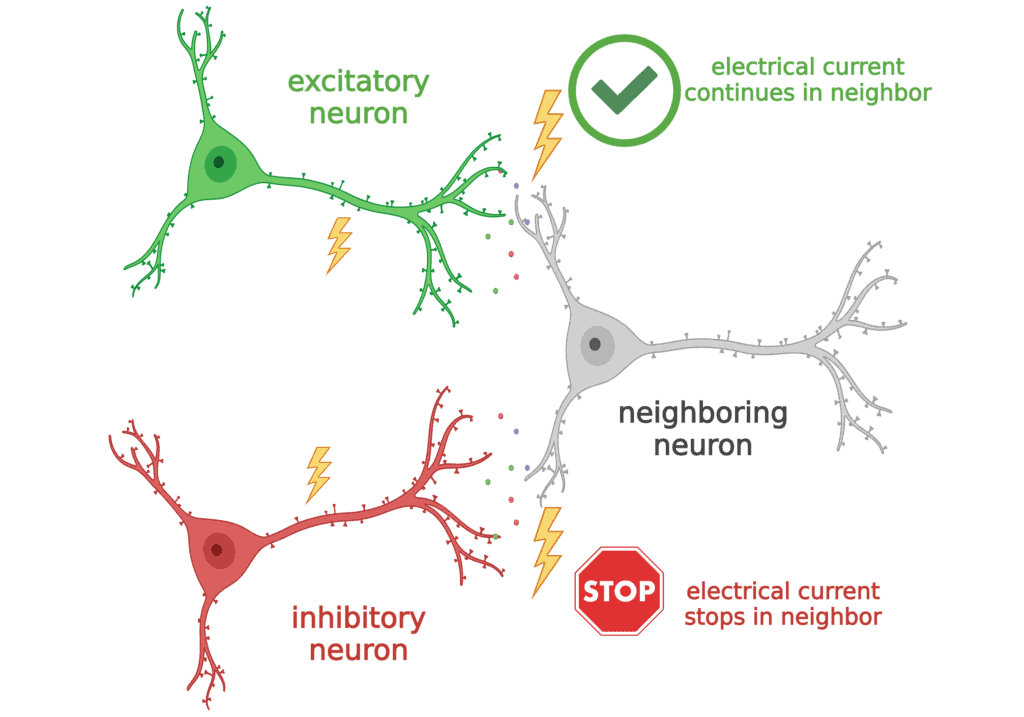
The sodium channel Nav1.1 is most important to inhibitory neurons. So in Dravet syndrome, these inhibitory neurons have trouble communicating. Normally, they should be telling the excitatory neurons when to STOP, but since they cannot do that properly, the excitatory neurons can get “too excited”, or generate too much electrical current, which leads to seizures.
Scientists are still learning a lot about how signaling in the brain is changed by the mutations that cause Dravet syndrome While disruptions to signaling from inhibitory neurons seems to be the main problem in Dravet syndrome, where essentially the brain is unable to “put the brakes” on electrical activity, there are also some other changes that impact other cell types. Additionally, there may be changes in brain activity that occur or change over time in response to the underlying genetic disruptions and ongoing seizure activity.




