Gene Therapy for Dravet Syndrome
Information regarding the state-of-the-field for disease-modifying therapeutic approaches.
Current Progress on a Gene Therapy for Dravet Syndrome
Last Updated August 2024
Is there a gene therapy for Dravet syndrome?
Several years ago, a genetic therapy still seemed a distant possibility for Dravet syndrome. However, advances to our understanding of genetics and medicine have allowed researchers to develop new approaches to gene-based interventions, bringing the possibility of disease-modifying therapies closer to reality for Dravet syndrome. Currently, there are two experimental therapies being tested in human clinical trials that target the genetic cause of Dravet syndrome. Additional approaches to overcome the genetic cause of Dravet syndrome are also in various stages of development.
What is the genetic cause of Dravet syndrome?
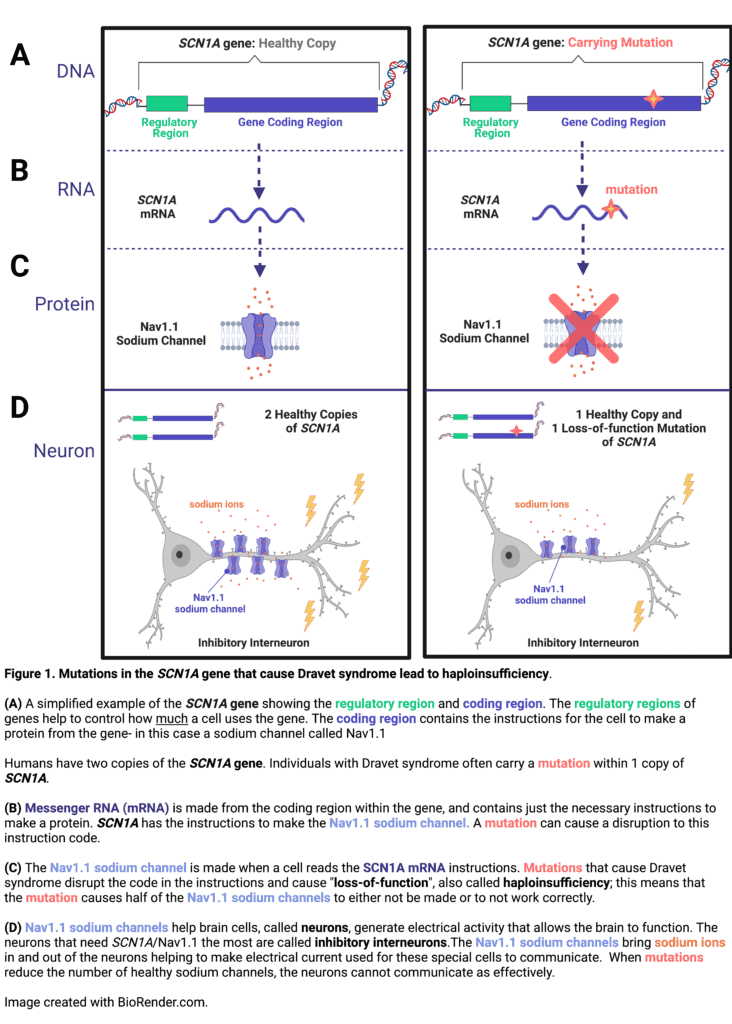
In over 90% of cases, Dravet syndrome is caused by a mutation in one copy of SCN1A, a gene that encodes a specific sodium channel, called Nav1.1, which is particularly important for some cells in the brain to communicate.1–3 Mutations in SCN1A that are associated with Dravet syndrome result in about 50% decreased expression or function of the Nav1.1 sodium channel (see Figure 1). This type of reduction in gene expression is referred to as a haploinsufficiency.4 One straightforward approach to a genetic therapy for a haploinsufficiency would be to deliver a replacement copy of the healthy gene. However, a major hurdle in the case of SCN1A is that the gene is very large and cannot fit inside the delivery vehicles (called vectors) that are currently utilized in genetic therapies. In addition to the difficulty of the gene size, a genetic therapy for Dravet syndrome would need to be delivered to cells in the brain, which can be more challenging to reach than some of the other organs in the body. Despite these challenges, researchers have discovered several creative solutions to overcome the genetic haploinsufficiency that causes Dravet syndrome.
Understanding DNA-RNA-Protein - A Recipe Analogy
Genetic-based therapies can be complex and understanding how they work can require some knowledge of basic molecular biology; mainly understanding DNA, genes, RNA, and protein. A good analogy to help explain these relationships is to think of a recipe blog or celebrity recipe book. DNA is the entire collection of recipes; genes are individual descriptions of recipes; RNA is the recipe card with just the important instructions; and protein is the completed recipe.
Many of us have come across recipes in blogs posts (or in elaborate cookbooks) that are full of extra tidbits about the background of a recipe with the actual recipe being almost hidden within the content. While this can be frustrating when you are just looking for the recipe, it’s actually a great analogy to the complexity of DNA!
Think of DNA as the entire catalogue of a cooking blog with lots of different recipes. Individual blog posts detailing a recipe represent unique genes. Cooking blog posts are often full of a lot of extra details that tell you all about the background of the recipe, when the author likes to make the recipe, how many servings the recipe will make, and perhaps even ways the recipe can be varied. DNA is actually very similar! There is a lot of information in the DNA surrounding the code for an individual gene that provides instructions for things like which cells should use the gene, when they should use the gene, how they might modify the gene for different situations, and how much of the gene to make.
RNA is represented by the actual core instructions for the recipe, which in this analogy is often buried within the entire recipe blog post. For practical purposes, it can be beneficial to just save or print the recipe card, rather than save the entire blog post. In the case of a cookbook, copying down the recipe ensures the cookbook stays clean and allows the recipe to be shared more easily. In just the same way, DNA is the master copy of gene recipes, with a lot of extra information around each gene. It is beneficial for the cells in your body to keep the DNA protected, while it uses RNA as ‘recipe cards’ to carry the streamlined instructions. Cells make a transcript from the DNA (in a process called transcription) that just contains the most important information for making the gene ‘recipe’. Cells follow instructions laid out in the DNA about where the gene “recipe” begins and ends and splice out information that is not necessary. Some genes use a process called alternative splicing, which allows a few variations on the recipe to be made from one gene; an analogy might be a recipe for bread where you swap add-ins like garlic or nuts to get a slightly varied end product. The RNA transcript, called messenger RNA (mRNA) then can be used by the cell as instructions to make the recipe.
Finally, the recipe instructions encoded in the RNA are used to make the actual gene recipe- a protein. This biological process is called translation, and would be equivalent to actually cooking the recipe. In the case of SCN1A, the mRNA contains the recipe to make a sodium channel called Nav1.1. This sodium channel is used primarily by inhibitory neurons in the brain that help to calm electrical activity used in neuronal communication (See Figure 1 for more details). When there is a genetic mutation in SCN1A, the recipe instructions may no longer make sense, either stopping too early before the recipe is complete, missing steps, or including incorrect steps.
Written by Veronica Hood, PhD; DSF Scientific Director
A variety of approaches are being developed to address Dravet syndrome at the genetic cause, hopefully going beyond symptomatic treatment and truly modifying the course of the disease.
I. Targeting DNA Regulation
When a gene within your DNA (like SCN1A) is expressed, the first step is to make a strand of messenger RNA (mRNA) that can then be used as instructions to make the final product: a protein (the sodium channel, Nav1.1). [If you need a quick overview on DNA-RNA-protein, check out the analogy above.] In order for a specific cell to know when and how much of a gene to express, there are regulatory regions in the DNA (often called promoters or enhancers) that act as markers to control gene expression. Molecules that bind to these regulatory regions and tune gene expression (on, off, up, or down) are called “transcription factors” (because the process of making mRNA from DNA is called “transcription”). Researchers have been working to identify where these regulatory regions are for SCN1A and ways that they can affect the regulatory regions to increase expression of SCN1A.
ETX101: A cell-selective gene regulation therapy delivered by AAV vector. An approach to increase expression of SCN1A called ETX101 is in development by Encoded Therapeutics. Instead of delivering a new copy of SCN1A, ETX101 delivers a regulatory gene that acts to specifically increase expression of SCN1A. The gene that ETX101 will be delivering to cells is called an engineered transcription factor. Since this engineered transcription factor is much smaller than the SCN1A gene, ETX101 can fit within a commonly used delivery packaging, called an adeno-associated viral (AAV) vector. ETX101 is anticipated to be a one-time-treatment delivered directly to the brain, where the engineered transcription factor will be expressed in the major type of neurons that use the SCN1A gene (a neuron type called GABA-ergic inhibitory interneurons). The hope is that this will restore the function of these neurons, reducing or eliminating seizures and other symptoms of Dravet syndrome. The preclinical work for ETX101 was published in 2022 and provided evidence that this approach could increase SCN1A expression, decrease seizures, and increase survival in a mouse model of Dravet syndrome. Additionally in non-human primates ETX101 was well-tolerated and delivered broadly across the brain.5 Encoded Therapeutics received approval to begin human studies of ETX101 in patients with Dravet syndrome in 2024, starting with small groups of patients in the United States, United Kingdom, and Australia. You can read more about the approvals and study opportunities in this blog post.
CRISPR. CRISPR technology has gained a lot of notoriety in recent years. Commonly, CRISPR technology is used to “cut and paste” sequences of DNA, and the therapeutic potential has largely focused on the ability to cut out and correct mutations in DNA and replace them with the correctg genetic information. Notably, this would still be a challenging approach for Dravet syndrome (at least with the current technology) given the size of the SCN1A gene and the large number of unique mutations that span across the entire SCN1A gene that are represented within the patient population. However, some researchers have been utilizing CRISPR technology in a different way to increase gene expression. Traditionally, ‘CRISPR associated protein 9’ or “Cas9”, is used in conjunction with a “guide sequence” to locate the target DNA sequence (usually around the spot where a gene mutation is present) and make a cut. Scientists have created a deactivated version of Cas9, called dCas9, that no longer harbors the ability to cut DNA, but instead can be connected to molecules that increase gene expression. This approach is also referred to as ‘CRISPR activation’ or ‘CRISPRa.’ Several groups of academic researchers are investigating how this technology could be utilized to increase SCN1A expression by targeting dCas9 to specific regulatory regions for SCN1A.6,7 Work in cell lines and mouse models of Dravet syndrome have shown the effectiveness of this approach to increase SCN1A, and consequently levels of the Nav1.1 sodium channel. Additionally, experiments indicate that this treatment approach can improve neuronal communication and seizure activity in Dravet syndrome mice. While encouraging, this work is still in preclinical development; there are challenges to the delivery method and efficiency of increasing gene expression. One of these studies used injection of multiple delivery vectors to contain both the dCas9 and the guide RNA sequences with limited expression in the brain, and the other study took advantage of mouse genetics to ensure the efficient delivery to the correct cells. Despite the need for those advancements to the technology for eventual human therapies, these proof-of-concept studies highlight that this approach could correct the haploinsufficiency of SCN1A and improve patient outcomes.
II. Targeting RNA Regulation
Regulation of expression can also occur at the RNA-level. Scientists are taking advantage of some of those regulatory instructions to increase the amount of Nav1.1 that SCN1A mRNA creates by targeting alternative splicing, correction of nonsense mutations, and stabilization of SCN1A mRNA transcripts.
TANGO ASO: Zorevunersen (STK-001) uses Targeted Augmentation of Nuclear Gene Output (TANGO) to Increase Expression of SCN1A with an Anti-sense Oligonucleotide (ASO). Stoke Therapeutics has developed a strategy called zorevunersen (STK-001) that works at the level of RNA-splicing. RNA-splicing occurs to remove the sections copied from the DNA code that are not essential to the “recipe” for creating the protein product (in this case, Nav1.1). The SCN1A mRNA sometimes includes a section of the DNA code called a poison exon within the RNA transcript that tells the cell to trash the strand of RNA instead of using it to produce the Nav1.1 protein. Stoke Therapeutics’ approach, called Target Augmentation of Nuclear Gene Output (TANGO), sends in a small piece of RNA that blocks the inclusion of the poison exon, and thus, increases the amount of RNA strands that produce Nav1.1.8 The small piece of RNA, called an antisense oligonucleotide or ASO, can enter cells and interact with targeted sections of genetic material. The advantage to this approach from Stoke is that only the cells that are naturally expressing SCN1A should be affected by the therapy, potentially helping to reduce off-target effects. Preclinical work showed efficacy to reduce seizures and mortality in mouse models of Dravet syndrome.9,10
Phase 1/2a clinical trials began in 2020 in the US and then UK for zorevunersen (STK-001) to determine the safety, pharmacokinetics, and efficacy in patients with Dravet syndrome. Zorevunersen is delivered by intrathecal injection (a similar process to a lumbar puncture or an epidural). Zorevunersen (STK-001) is not a permanent or one-time therapy; instead, zorevunersen will likely need to be administered every four months. The recently completed Phase 1/2a trials reported significant reductions in seizure frequency and patients that have transitioned to continued dosing in the open-label extension studies have shown significant improvements in measures of cognition and behavior. This data suggests zorevunersen (STK-001) may be the first ever disease-modifying therapy for Dravet syndrome (read more in this blog). Stoke Therapeutics is beginning a global Phase 3 study in 2025 to continue to assess the safety and efficacy of zorevunersen in patients with Dravet syndrome.
tRNA. Tevard Biosciences (who partnered with Zogenix, now a part of UCB in late 2020) is working to advance two therapies that could correct the SCN1A haploinsufficiency in Dravet syndrome. They use a different kind of RNA, called transfer RNA or tRNA, to increase SCN1A expression. The first approach specifically targets nonsense mutations. Nonsense mutations create a change in the DNA code that tells the cell to prematurely stop making the Nav1.1 protein, leading to a shortened version that gets broken down by the cell (or does not work as efficiently as it should). The therapy in development uses a tRNA that can overcome the nonsense mutation by reading through the premature stop signal and allowing the Nav1.1 protein to be made correctly.11 The other therapy they are developing also uses tRNA, but this approach helps to stabilize the SCN1A mRNA so that it can be used to create more copies of the Nav1.1 protein. Both of these approaches would be delivered in an AAV vector to cells in the brain, making them a one-time therapy. In addition to these tRNA approaches, in the spring of 2022, Tevard also added a third approach to their developmental pipeline, called an mRNA-amplifier, to increase expression from mRNA strands.12 Currently, the preclinical experiments have all been done in cells, and the company is now working with animal models of Dravet syndrome. These therapies are exciting because of the potential they hold for broad application to other genes, but there is still a lot of proof-of-concept work that will have to be done.
III. Focusing on Alternative Approaches to Gene Replacement
As mentioned above, one of the major challenges to overcome for gene therapy in Dravet syndrome has been the size of SCN1A, which is too large to fit into the vectors that would be most ideal for delivery to neurons in the brain. Several academic research collaboratives are working on utilizing larger types of adenoviral vectors to deliver a full replacement copy of SCN1A.14,15 Another method being investigated is if the SCN1A gene could be split in half, delivered in two smaller AAV delivery vectors and then the two parts of the gene reassemble in the target cell.16 All of these studies are still in early stages, but the work in cells and mouse models have promising results. It is yet to be determined what the challenges of using these gene replacement approaches in humans might be, but the field is marching forward steadily.
IV. Targeting Genes that Can Compensate for SCN1A Haploinsufficiency
A few researchers have attempted to compensate for the decreased expression of SCN1A by altering other genes in the same pathway. One group sent in another copy of a gene (NaV1B) that interacts with the sodium channel Nav1.1, with mixed results based on sex in a mouse model of Dravet syndrome and overall modest impacts on seizures.17 Another group has decreased expression of a separate sodium channel gene, SCN8A, which eliminated behavioral (motor) seizures (for 5 months post-treatment) and reduced mortality in a mouse model of Dravet syndrome.18 In 2021, DSF awarded a post-doctoral fellowship to a researcher working on this approach.
In summary, there are several disease-modifying therapeutic approaches in various stages of development for Dravet syndrome. It is encouraging to see so many different tactics being employed to overcome the challenges to correcting the haploinsufficiency of SCN1A in Dravet syndrome. DSF has worked diligently to fund basic research that lays the groundwork for the development of genetic therapies. DSF also continues to work closely with industry as they develop therapies for human patients.
Learn more about the genetics of Dravet syndrome.
Stay up-to-date on enrolling clinical studies and the therapeutic pipeline for Dravet syndrome.
More questions about this topic? Email DSF’s Scientific Director, Veronica Hood, PhD: veronica@dravetfoundation.org
References:
Zuberi SM, Brunklaus A, Birch R, Reavey E, Duncan J, Forbes GH. Genotype–phenotype associations in SCN1A-related epilepsies. Neurology. 2011;76(7):594-600. doi:10.1212/wnl.0b013e31820c309b
Wu YW, Sullivan J, McDaniel SS, et al. Incidence of Dravet Syndrome in a US Population. Pediatrics. 2015;136(5):e1310-e1315. doi:10.1542/peds.2015-1807
Li W, Schneider AL, Scheffer IE. Defining Dravet syndrome: An essential pre‐requisite for precision medicine trials. Epilepsia. Published online 2021. doi:10.1111/epi.17015
Catterall WA, Kalume F, Oakley JC. NaV1.1 channels and epilepsy. J Physiology. 2010;588(11):1849-1859. doi:10.1113/jphysiol.2010.187484
Tanenhaus A, Stowe T, Young A, et al. Cell-Selective Adeno-Associated Virus-Mediated SCN1A Gene Regulation Therapy Rescues Mortality and Seizure Phenotypes in a Dravet Syndrome Mouse Model and Is Well Tolerated in Nonhuman Primates. Hum Gene Ther. 2022;33(11-12):579-597. doi:10.1089/hum.2022.037
Colasante G, Lignani G, Brusco S, et al. dCas9-Based Scn1a Gene Activation Restores Inhibitory Interneuron Excitability and Attenuates Seizures in Dravet Syndrome Mice. Mol Ther. 2020;28(1):235-253. doi:10.1016/j.ymthe.2019.08.018
Yamagata T, Raveau M, Kobayashi K, et al. CRISPR/dCas9-based Scn1a gene activation in inhibitory neurons ameliorates epileptic and behavioral phenotypes of Dravet syndrome model mice. Neurobiol Dis. 2020;141:104954. doi:10.1016/j.nbd.2020.104954
Lim KH, Han Z, Jeon HY, et al. Antisense oligonucleotide modulation of non-productive alternative splicing upregulates gene expression. Nat Commun. 2020;11(1):3501. doi:10.1038/s41467-020-17093-9
Han Z, Chen C, Christiansen A, et al. Antisense oligonucleotides increase Scn1a expression and reduce seizures and SUDEP incidence in a mouse model of Dravet syndrome. Sci Transl Med. 2020;12(558):eaaz6100. doi:10.1126/scitranslmed.aaz6100
Wengert ER, Wagley PK, Strohm SM, et al. Targeted Augmentation of Nuclear Gene Output (TANGO) of SCN1A Rescues Parvalbumin Interneuron Excitability and Reduces Seizures in a Mouse Model of Dravet Syndrome. Brain Res. 2021;1775:147743. doi:10.1016/j.brainres.2021.147743
Lueck JD, Yoon JS, Perales-Puchalt A, et al. Engineered transfer RNAs for suppression of premature termination codons. Nat Commun. 2019;10(1):822. doi:10.1038/s41467-019-08329-4
Torkzaban B, Kawalerski R, Coller J. Development of a Tethered mRNA Amplifier to increase protein expression. Biotechnol J. Published online 2022:2200214. doi:10.1002/biot.202200214
Hsiao, J., Yuan, T.Y., Tsai, M.S., Lu, C.Y., Lin, Y.C., Lee, M.L., Lin, S.W., Chang, F.C., Pimentel, H.L., Olive, C., Coito, C., Shen, G., Young, M., Thorne, T., Lawrence, M., Magistri, M., Faghihi, M.A., Khorkova, O., Wahlestedt, C., 2016. Upregulation of Haploinsufficient Gene Expression in the Brain by Targeting a Long Non-coding RNA Improves Seizure Phenotype in a Model of Dravet Syndrome. Ebiomedicine 9, 257–277. doi:10.1016/j.ebiom.2016.05.011
Mora-Jimenez L, Valencia M, Sanchez-Carpintero R, et al. Transfer of SCN1A to the brain of adolescent mouse model of Dravet syndrome improves epileptic, motor, and behavioral manifestations. Mol Ther – Nucleic Acids. 2021;25:585-602. doi:10.1016/j.omtn.2021.08.003
Fadila S, Beucher B, Reyes IGD, et al. Exogenous NaV1.1 activity in excitatory and inhibitory neurons reverts Dravet syndrome comorbidities when delivered post-symptom onset in mice with Dravet. Biorxiv. Published online 2022:2022.06.10.495591. doi:10.1101/2022.06.10.495591
Mich JK, Ryu J, Wei AD, et al. AAV-mediated interneuron-specific gene replacement for Dravet syndrome. BioRx Preprint 2023; doi: 10.1101/2023.12.15.571820
Niibori Y, Lee SJ, Minassian BA, Hampson DR. Sexually Divergent Mortality and Partial Phenotypic Rescue After Gene Therapy in a Mouse Model of Dravet Syndrome. Hum Gene Ther. 2020;31(5-6):339-351. doi:10.1089/hum.2019.225
Lenk GM, Jafar‐Nejad P, Hill SF, et al. Scn8a Antisense Oligonucleotide Is Protective in Mouse Models of SCN8A Encephalopathy and Dravet Syndrome. Ann Neurol. 2020;87(3):339-346. doi:10.1002/ana.25676





